Setting up custom alerts
custom-alerts.Rmd
library(slurmtools)
#> Error in get(paste0(generic, ".", class), envir = get_method_env()) :
#> object 'type_sum.accel' not found
#>
#>
#> ── Needed slurmtools options ───────────────────────────────────────────────────
#> ✖ options('slurmtools.slurm_job_template_path') is not set.
#> ✖ options('slurmtools.submission_root') is not set.
#> ✖ options('slurmtools.bbi_config_path') is not set.
#> ℹ Please set all options for job submission defaults to work.
library(bbr)
library(here)
#> here() starts at /home/runner/work/slurmtools/slurmtools
nonmem = file.path(here::here(), "vignettes", "model", "nonmem")
options('slurmtools.submission_root' = file.path(nonmem, "submission-log"))Submitting a NONMEM job with nmm
Instead of using bbi we can use nmm (NONMEM
Monitor) which currently has some additional functionality of
sending notifications about zero gradients, missing -1E9 lines in ext
file, and some very basic control stream errors. Nonmem-monitor also
allows for setting up an alerter to be better fed these messages - more
on that later. To use nmm you can install the latest
release from the github repository linked above.
We can update the template file accordingly:
#!/bin/bash
#SBATCH --job-name="{{job_name}}"
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task={{ncpu}}
#SBATCH --partition={{partition}}
{{nmm_exe_path}} -c {{config_toml_path}} rundefault, submit_slurm_job will provide
nmm_exe_path and config_toml_path to the
template. Just like with bbi_exe_path,
nmm_exe_path is determined with
Sys.which("nmm") which may or may not give you the path to
the nmm binary if it is on your path or not. We can inject the
nmm_exe_path like we did with bbi_exe_path and
assume it’s not on our path.
The config.toml file controls what nmm will
monitor and where to look for files and how to alert you. We’ll use
generate_nmm_config() to create this file. First we can
look at the documentation to see what type of information we should pass
to this function. ?generate_nmm_config()
mod_number <- "1001"
if (file.exists(file.path(nonmem, paste0(mod_number, ".yaml")))) {
mod <- bbr::read_model(file.path(nonmem, mod_number))
} else {
mod <- bbr::new_model(file.path(nonmem, mod_number))
}
slurmtools::generate_nmm_config(mod)
#> Warning in slurmtools::generate_nmm_config(mod): bbi.yaml path not supplied
#> through options('slurmtools.bbi_config_path') or through argumentsThis generates the following toml file. By passing in just the mod
object, nmm will use the default values for the other
options so if you need to change which files are tracked, or how many
threads to use you’ll have to explicitly pass that to
generate_nmm_config. Since we’re in vignettes we’ll need to
update the watched_dir and output_dir
accordingly.
model_number = '1001'
watched_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/model/nonmem'
output_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/model/nonmem/in_progress'
slurmtools::generate_nmm_config(
mod,
watched_dir = "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem",
output_dir = "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/in_progress",
bbi_config_path = "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/bbi.yaml"
)This updates the 1001.toml config file to:
model_number = '1001'
watched_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem'
output_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/in_progress'We can now run submit_slurm_job and get essentially the
same behavior as running with bbi. On linux
~/.local/bin/ will be on your path so saving the downloaded
binaries there is a good approach.
submission_nmm <- slurmtools::submit_slurm_job(
mod,
overwrite = TRUE,
slurm_job_template_path = file.path(nonmem, "slurm-job-nmm.tmpl"),
slurm_template_opts = list(
nmm_exe_path = normalizePath("~/.local/bin/nmm"))
)
#> Warning in normalizePath("~/.local/bin/nmm"):
#> path[1]="/home/runner/.local/bin/nmm": No such file or directory
submission_nmm
#> $status
#> [1] 0
#>
#> $stdout
#> [1] "Submitted batch job 804\n"
#>
#> $stderr
#> [1] ""
#>
#> $timeout
#> [1] FALSE
slurmtools::get_slurm_jobs(user = "matthews")
#> # A tibble: 1 × 13
#> job_id partition job_name user_name job_state time cpus standard_input
#> <int> <chr> <chr> <chr> <chr> <time> <int> <chr>
#> 1 1159 cpu2mem4gb 1001-nonmem… matthews COMPLETED 00'13" 1 /dev/null
#> # ℹ 5 more variables: standard_output <chr>, submit_time <dttm>,
#> # start_time <dttm>, end_time <dttm>, current_working_directory <chr>The one difference between using nmm compared to
bbi is that a new directory is created that contains a log
file that caught some issues with our run. This file is updated as
nonmem is running and monitors gradient values, parameters that hit
zero, as well as other errors from bbi. Looking at the first few lines
we can see that bbi was successfully able to call nonmem.
We also see an info level log that OMEGA(2,1) has 0 value – in our mod
file we don’t specify any omega values off the diagonal so these are
fixed at 0. Finally we see that GRD(6) hit 0 relatively early in the
run.
19:13:45 [INFO] bbi log: time="2024-09-20T19:13:45Z" level=info msg="Successfully loaded default configuration from /cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/bbi.yaml"
19:13:45 [INFO] bbi log: time="2024-09-20T19:13:45Z" level=info msg="Beginning Local Path"
19:13:45 [INFO] bbi log: time="2024-09-20T19:13:45Z" level=info msg="A total of 1 models have completed the initial preparation phase"
19:13:45 [INFO] bbi log: time="2024-09-20T19:13:45Z" level=info msg="[1001] Beginning local work phase"
19:14:16 [INFO] "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001/1001.ext": Iteration: 5, Parameter(s) that hit zero: ["SIGMA(2,1)", "OMEGA(2,1)"]
19:14:19 [INFO] "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001/1001.ext": Iteration: 10, Parameter(s) that hit zero: ["OMEGA(2,1)", "SIGMA(2,1)"]
19:14:21 [INFO] "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001/1001.ext": Iteration: 15, Parameter(s) that hit zero: ["SIGMA(2,1)", "OMEGA(2,1)"]
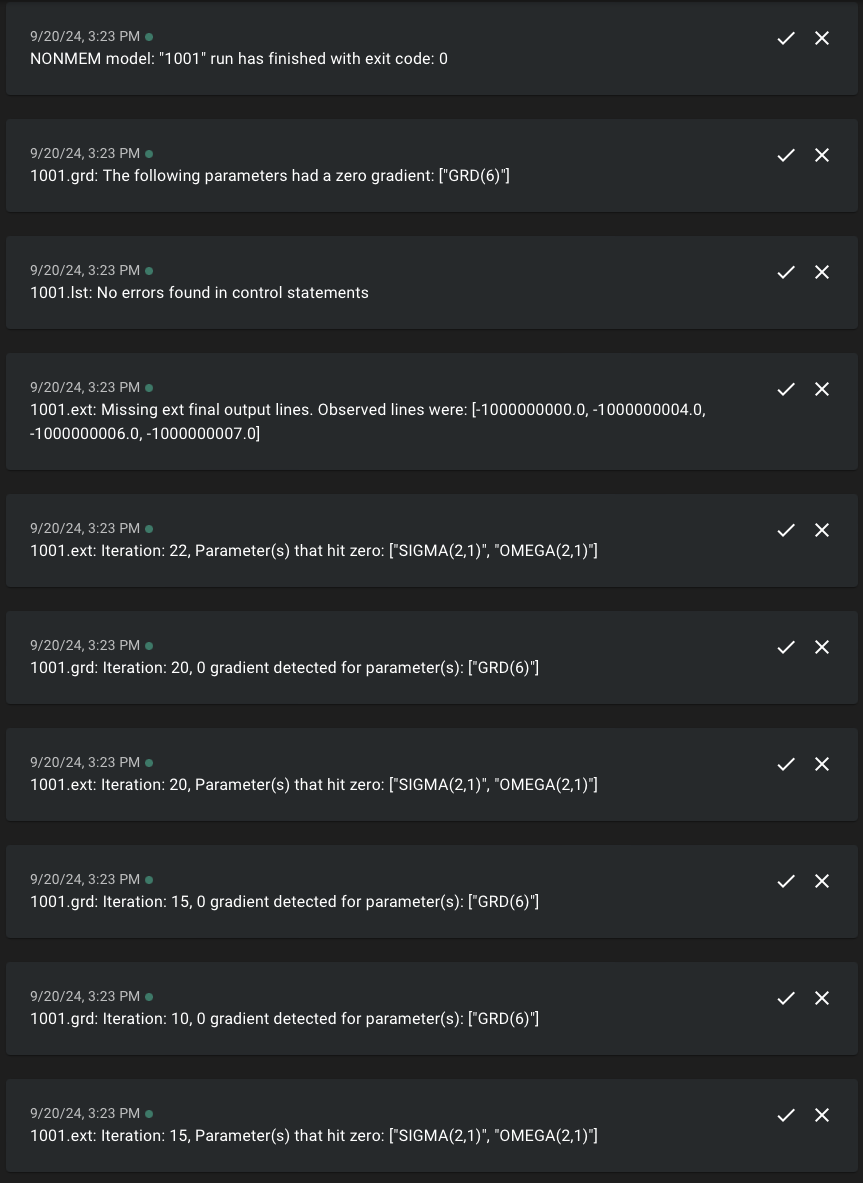
19:14:21 [WARN] "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001/1001.grd" Iteration: 10, has 0 gradient for parameter(s): ["GRD(6)"] After a run has finished several messages are sent to the log after a
final check of the files listed in the files_to_track field
of the 1001.toml file.
19:14:31 [INFO] Received Exit code: exit status: 0
19:14:31 [WARN] 1001.ext: Missing ext final output lines. Observed lines were: [-1000000000.0, -1000000004.0, -1000000006.0, -1000000007.0]
19:14:31 [WARN] "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001/1001.grd": The following parameters hit zero gradient through the run: ["GRD(6)"]We see that GRD(6) hit zero during the run and that only a subset of the -1E9 lines were present in the .ext file.
Getting alerted during a run
Like we did with bbi and altering the slurm template
file to get notifications from ntfy.sh
nmm has this feature built in! The messages in the log file
that relate to zero gradients, missing -1E9 lines, and 0 parameter
values can also be sent to ntfy by altering the 1001.toml
file. We can get these alerts in real time without having to dig through
a noisy log file.
Let’s update our call to generate_nmm_config to have
nmm send notifications to the NONMEMmonitor
topic on ntfy.sh. Just like how
submit_slurm_job can feed additional information to the
template with slurm_template_opts, we can add an alerter
feature to nmm with alerter_opts. If we go to ntfy.sh we
can see that to send a message to ntfy we can run
curl -d "Backup successful 😀" ntfy.sh/mytopic.
nmm can call a binary with a command and pass a message to
a flag. For ntfy, the binary is curl the message flag is
d and the command is ntfy.sh/mytopic and there
are no additional args.
slurmtools::generate_nmm_config(
mod,
watched_dir = "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem",
output_dir = "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/in_progress",
alerter_opts = list(
alerter = Sys.which('curl'), #binary location of curl,
command = "ntfy.sh/NONMEMmonitor",
message_flag = "d"
)
)
#> Warning in slurmtools::generate_nmm_config(mod, watched_dir =
#> "/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem",
#> : bbi.yaml path not supplied through options('slurmtools.bbi_config_path') or
#> through argumentsThis updates the 1001.toml file to this:
model_number = '1001'
watched_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem'
output_dir = '/cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/in_progress'
[alerter]
alerter = '/usr/bin/curl'
command = 'ntfy.sh/NONMEMmonitor'
message_flag = 'd'When we re-run the submit_slurm_job call we will now get
ntfy notifications. One thing to note is that nmm will
print full paths in the log, but will only send notifications with the
model_number (or
model_number.file_extension).
submission_nmm <- slurmtools::submit_slurm_job(
mod,
overwrite = TRUE,
slurm_job_template_path = file.path(nonmem, "slurm-job-nmm.tmpl"),
slurm_template_opts = list(
nmm_exe_path = normalizePath("~/.local/bin/nmm"))
)
#> Warning in normalizePath("~/.local/bin/nmm"):
#> path[1]="/home/runner/.local/bin/nmm": No such file or directory
submission_nmm
#> $status
#> [1] 0
#>
#> $stdout
#> [1] "Submitted batch job 804\n"
#>
#> $stderr
#> [1] ""
#>
#> $timeout
#> [1] FALSE
slurmtools::get_slurm_jobs(user = "matthews")
#> # A tibble: 1 × 13
#> job_id partition job_name user_name job_state time cpus standard_input
#> <int> <chr> <chr> <chr> <chr> <time> <int> <chr>
#> 1 1159 cpu2mem4gb 1001-nonmem… matthews COMPLETED 00'13" 1 /dev/null
#> # ℹ 5 more variables: standard_output <chr>, submit_time <dttm>,
#> # start_time <dttm>, end_time <dttm>, current_working_directory <chr>This gives us the notifications in a much more digestible format