Running Nonmem with slurmtools
Running-nonmem.RmdSlurmtools for submitting NONMEM runs
slurmtools is an R package for interacting with slurm
(fka Simple Linux
Utility for Resource
Management) and submitting NONMEM jobs. You can submit
a NONMEM job with submit_slurm_job, you can view current
jobs with get_slurm_jobs, and you can see the available
partitions with get_slurm_partitions.
Installing slurmtools
To install slurmtools use the following commands:
library(slurmtools)
#> Error in get(paste0(generic, ".", class), envir = get_method_env()) :
#> object 'type_sum.accel' not found
#>
#>
#> ── Needed slurmtools options ───────────────────────────────────────────────────
#> ✖ options('slurmtools.slurm_job_template_path') is not set.
#> ✖ options('slurmtools.submission_root') is not set.
#> ✖ options('slurmtools.bbi_config_path') is not set.
#> ℹ Please set all options for job submission defaults to work.We are given a message when loading slurmtools that some options are
not set and that default job submission will not work without them.
These options are used for default arguments in the
submit_slurm_job function. Running
?submit_slurm_job we can see the documentation
Default variables and values provided to the template
This function uses the inputs to populate a template Bash shell script that submits the NONMEM job to slurm. A default template file is supplied with the Project Starter and it can be modified to do additional tasks as long as they are possible within Bash.
By default these values are provided to the slurm template file:
partitionis an argument tosubmit_slurm_jobby default this will be the first element ofget_slurm_partitions()parallelisTRUEifncpu > 1, elseFALSEncpuis an argument tosubmit_slurm_job- your input ofncpumust be less than or equal to the number of cpus on thepartitionjob_nameis created from the.modargument supplied tosubmit_slurm_job- default is<nonmem_model_name>-nonmem-runproject_pathis determined fromhere::here()and should be the root of the Rprojectproject_nameis determined fromhere::here() %>% basename()- for this project it would beslurmtoolsbbi_exe_pathis determined via `Sys.which(“bbi”)bbi_config_pathis determined via getOption(“slurmtools.bbi_config_path”)model_pathis determined from the.modargument supplied tosubmit_slurm_jobconfig_toml_pathis determined from the.modargument supplied tosubmit_slurm_joband is required to usenmm(NONMEM monitor)nmm_exe_pathis determined viaSys.which("nmm")
If you need to feed more arguments to the template you simply supply
them in the slurm_template_opts argument as a list. More on
that later.
default_template_list = list(
partition = partition,
parallel = parallel,
ncpu = ncpu,
job_name = sprintf("%s-nonmem-run", basename(.mod$absolute_model_path)),
project_path = project_path,
project_name = project_name,
bbi_exe_path = Sys.which("bbi"),
bbi_config_path = bbi_config_path,
model_path = .mod$absolute_model_path,
config_toml_path = config_toml_path,
nmm_exe_path = Sys.which("nmm")
)Submitting a NONMEM job with bbi
To submit a NONMEM job, we need to supply either the path to a mod
file or create a model object from bbr, and supply a
slurm-template.tmpl file. To use bbi we also
need a bbi.yaml file, which I’ve also supplied in
/model/nonmem/bbi.yaml (and is also supplied with the R
project starter).
Here is an example of a template file that will call
bbi:
#!/bin/bash
#SBATCH --job-name="{{job_name}}"
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task={{ncpu}}
#SBATCH --partition={{partition}}
#SBATCH --account={{project_name}}
#{{project_path}}
# submit_slurm_job uses the whisker package to populate template files
# https://github.com/edwindj/whisker
{{#parallel}}
{{bbi_exe_path}} nonmem run local {{model_path}}.mod --parallel --threads={{ncpu}} --config {{bbi_config_path}}
{{/parallel}}
{{^parallel}}
{{bbi_exe_path}} nonmem run local {{model_path}}.mod --config {{bbi_config_path}}
{{/parallel}}This file will call bbi to run our supplied model
({{model_path}}.mod) if ncpu > 1 then
parallel will be true and the code between {{#parallel}}
and {{/parallel}} will be populated. if
ncpu = 1 then parallel will be false and the code between
{{^parallel}} and {{/parallel}} will be
populated. By default, submit_slurm_job will inject
Sys.which("bbi") into the template, so if bbi
is not on your path we’ll have to supply the bbi_exe_path
for it to start the NONMEM run.
Sys.which("bbi")
#> bbi
#> ""We will use a few different template files with different
functionality so we’ll inject those template file paths to
submit_slurm_job. However, we’ll use the
submission-log directory for the output, so we’ll set that
option as well as bbi_config_path so
submit_slurm_job defaults can be used. The slurm template
files are saved in ~/model/nonmem/ Additionally, there is a
simple NONMEM control stream in 1001.mod in the same
directory that we can use for testing.
library(bbr)
library(here)
#> here() starts at /home/runner/work/slurmtools/slurmtools
nonmem = file.path(here::here(), "vignettes", "model", "nonmem")
options('slurmtools.submission_root' = file.path(nonmem, "submission-log"))
options('slurmtools.bbi_config_path' = file.path(nonmem, "bbi.yaml"))To create the bbr model object, we need to have both
1001.mod and 1001.yaml which contains metadata
about the model in the supplied directory
(./model/nonmem/). We’ll check for mod_number.yaml and if
it exists, read in the model otherwise create it and then read it.
mod_number <- "1001"
if (file.exists(file.path(nonmem, paste0(mod_number, ".yaml")))) {
mod <- bbr::read_model(file.path(nonmem, mod_number))
} else {
mod <- bbr::new_model(file.path(nonmem, mod_number))
}We can now submit the job and point to the template file in
model/nonmem/slurm-job-bbi.tmpl.
submission <- slurmtools::submit_slurm_job(
mod,
overwrite = TRUE,
slurm_job_template_path = file.path(nonmem, "slurm-job-bbi.tmpl"),
)
submission
#> $status
#> [1] 0
#>
#> $stdout
#> [1] "Submitted batch job 804\n"
#>
#> $stderr
#> [1] ""
#>
#> $timeout
#> [1] FALSEWe see a status with an exit code of 0 suggesting a
successful command, and the stdout gives us the batch job
number. We can use slurmtools::get_slurm_jobs() to monitor
the status of the job. Here, we can supply the user = “matthews”
argument to filter to just the jobs I’ve submitted.
slurmtools::get_slurm_jobs(user = "matthews")
#> # A tibble: 1 × 13
#> job_id partition job_name user_name job_state time cpus standard_input
#> <int> <chr> <chr> <chr> <chr> <time> <int> <chr>
#> 1 1159 cpu2mem4gb 1001-nonmem… matthews COMPLETED 00'13" 1 /dev/null
#> # ℹ 5 more variables: standard_output <chr>, submit_time <dttm>,
#> # start_time <dttm>, end_time <dttm>, current_working_directory <chr>If we look in the slurmtools.submisstion_root directory
we can see the shell script that was generated with
submit_slurm_job. Here is the whisker replaced call to
bbi:
#!/bin/bash
#SBATCH --job-name="1001-nonmem-run"
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --partition=cpu2mem4gb
#SBATCH --account=slurmtools
#/cluster-data/user-homes/matthews/Packages/slurmtools
# submit_slurm_job uses the whisker package to populate template files
# https://github.com/edwindj/whisker
/usr/local/bin/bbi nonmem run local /cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/1001.mod --config /cluster-data/user-homes/matthews/Packages/slurmtools/vignettes/model/nonmem/bbi.yamlExtending templates
Because the templates create a bash shell script there is an almost infinite number of things we can do with our template. Anything you can do in bash you can do by appropriately updating the template file and injecting the needed information!
Let’s add a notification feature that will send a notification when the job has started and finished. We can use ntfy.sh and add the necessary info to our template to achieve this.
Here is a modified template file that adds a
JOBID=$SLURM_JOBID and some ntfy calls. To get a
notification we can supply submit_slurm_job with
ntfy variable to send notifications. I’ll use
ntfy = ntfy_demo for this.
#!/bin/bash
#SBATCH --job-name="{{job_name}}"
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task={{ncpu}}
#SBATCH --partition={{partition}}
#SBATCH --account={{project_name}}
JOBID=$SLURM_JOBID
# submit_slurm_job uses the whisker package to populate template files
# https://github.com/edwindj/whisker
{{#ntfy}}
curl -d "Starting model run: {{job_name}} $JOBID" ntfy.sh/{{ntfy}}
{{/ntfy}}
{{#parallel}}
{{bbi_exe_path}} nonmem run local {{model_path}}.mod --parallel --threads={{ncpu}} --config {{bbi_config_path}}
{{/parallel}}
{{^parallel}}
{{bbi_exe_path}} nonmem run local {{model_path}}.mod --config {{bbi_config_path}}
{{/parallel}}
{{#ntfy}}
curl -d "Finished model run: {{job_name}} $JOBID" ntfy.sh/{{ntfy}}
{{/ntfy}}Since we’ve already run this model we will provide the
overwrite = TRUE argument to force a new nonmem run.
submission_ntfy <- slurmtools::submit_slurm_job(
mod,
slurm_job_template_path = file.path(nonmem, "slurm-job-bbi-ntfy.tmpl"),
overwrite = TRUE,
slurm_template_opts = list(
ntfy = "ntfy_demo")
)
submission_ntfy
#> $status
#> [1] 0
#>
#> $stdout
#> [1] "Submitted batch job 804\n"
#>
#> $stderr
#> [1] ""
#>
#> $timeout
#> [1] FALSEWe again get a 0 exit code status and now instead of using
slurmtools::get_slurm_jobs() to monitor the job, we can
rely on the new notifications we just set up. 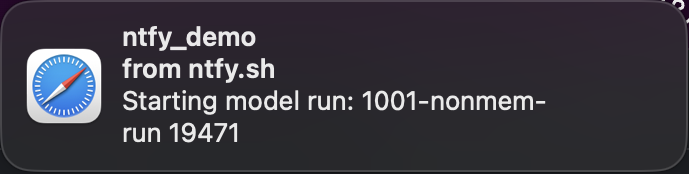
and when the run finished we get another notification: 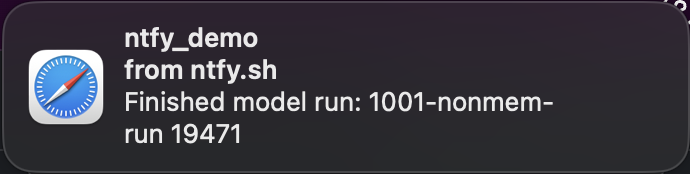
Note that the run number will match the run specified in
submission$stdout. We can see the new shell script this
updated template file generated
#!/bin/bash
#SBATCH --job-name="1001-nonmem-run"
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --partition=cpu2mem4gb
#SBATCH --account=slurmtools
JOBID=$SLURM_JOBID
curl -d "Starting model run: 1001-nonmem-run $JOBID" ntfy.sh/ntfy_demo
/usr/local/bin/bbi nonmem run local /cluster-data/user-homes/matthews/Projects/slurmtools_vignette/model/nonmem/1001.mod --config /cluster-data/user-homes/matthews/Projects/slurmtools_vignette/model/nonmem/bbi.yaml
curl -d "Finished model run: 1001-nonmem-run $JOBID" ntfy.sh/ntfy_demoTo reiterate, this template file is run as a bash shell script so anything you can do in bash you can put into the template and pass the needed arguments and customize the behavior to your liking.