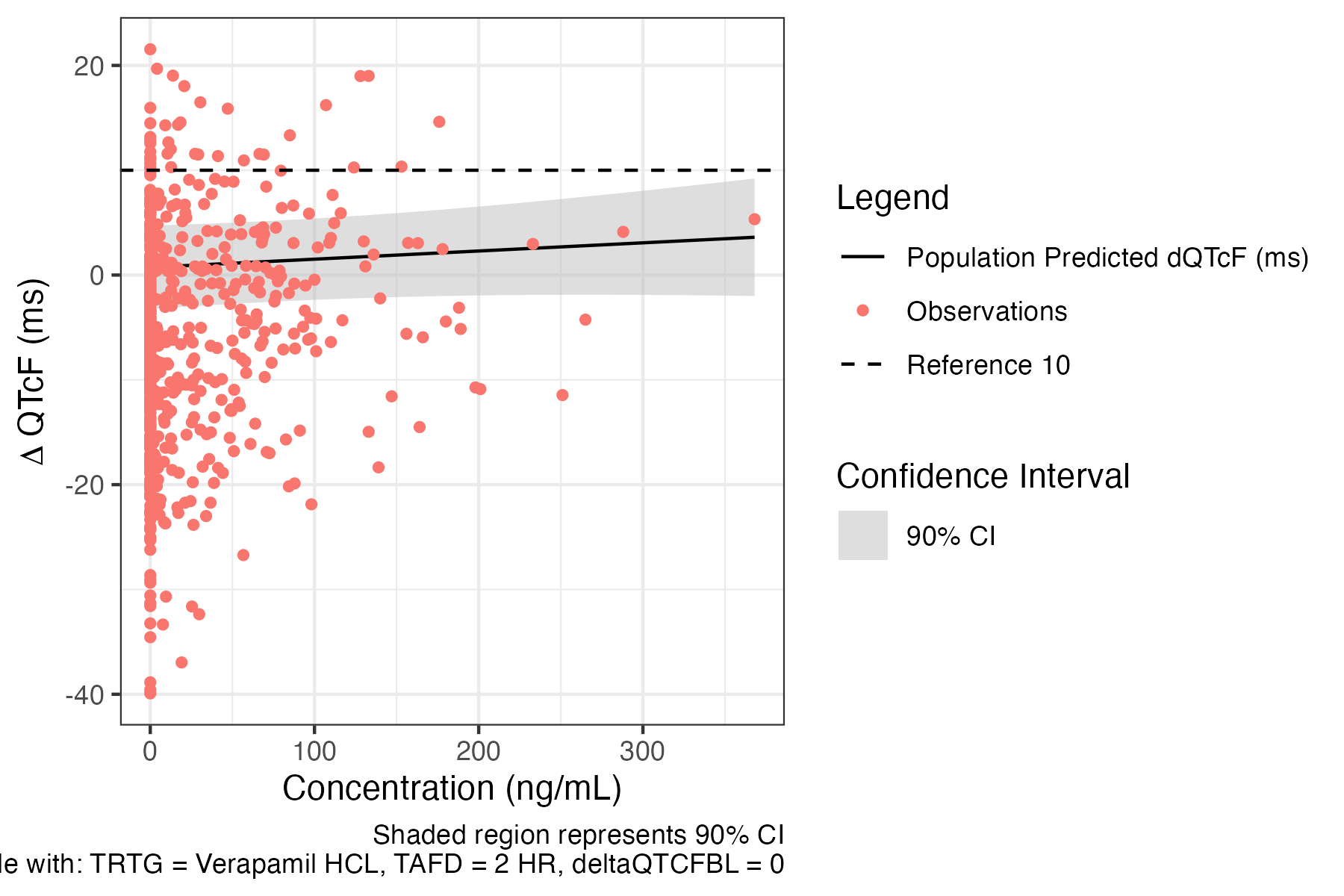
predict_with_observations_plot
Description
Plots predictions of the model with observed values
Usage
predict_with_observations_plot( data, fit, conc_col, dv_col, id_col = NULL, ntime_col = NULL, trt_col = NULL, treatment_predictors, control_predictors = NULL, reference_threshold = c( 10), conf_int = 0.9, contrast_method = c( "matched", "group"), style = list())Arguments
| Name | Description |
|---|---|
data | a dataframe of QTc dataset |
fit | the lme model to make predictions with |
conc_col | an unquoted column name of drug concentration measurements |
dv_col | an unquoted column name of dQTC measurements |
id_col | an unquoted column name of ID data, used when control predictors is provided to compute delta delta dv |
ntime_col | an unquoted column name of Nominal time data, used when control predictors is provided to compute delta delta dv |
trt_col | an unquoted column name of Treatment group data, used when control predictors is provided to compute delta delta dv |
treatment_predictors | a list for predictions with model. Should contain a value for each predictor in the model. |
control_predictors | an optional list for contrast predictions |
reference_threshold | optional vector of numbers to add as horizontal dashed lines |
conf_int | a float for the fractional confidence interval. default = 0.9 |
contrast_method | a string specifying contrast method when using control_predictors: “matched” for individual ID+time matching (crossover studies), “group” for group-wise subtraction (parallel studies) |
style | a named list of any argument that can be passed to style_plot |
Returns
a plot
Examples
data <- preprocess(data) fit <- fit_prespecified_model( data, deltaQTCF, ID, CONC, deltaQTCFBL, TRTG, TAFD, "REML", TRUE ) predict_with_observations_plot( data, fit, CONC, deltaQTCF, treatment_predictors = list( CONC = 0, TRTG = "Verapamil HCL", TAFD = "2 HR", deltaQTCFBL = 0 ), conf_int = 0.9 )