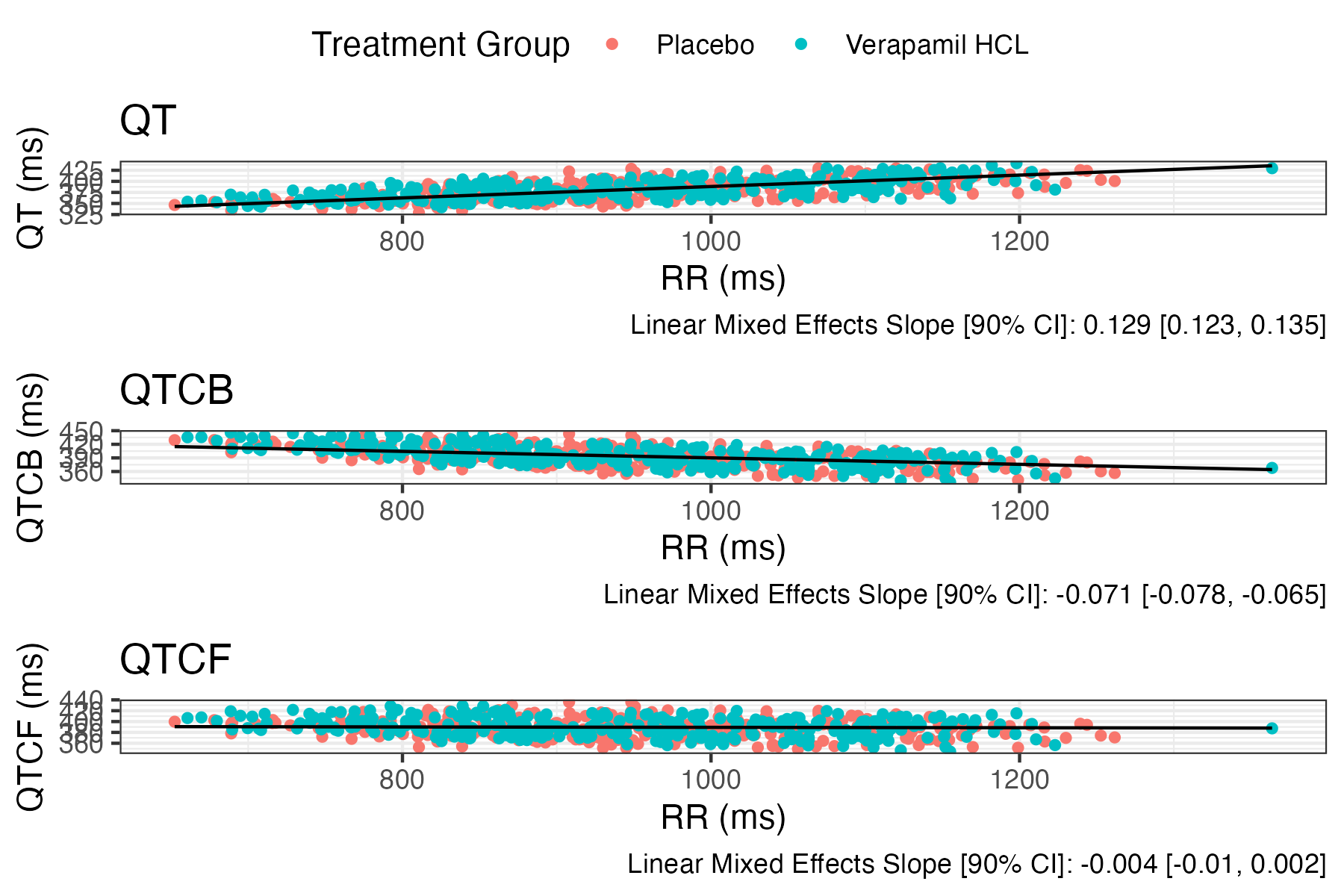
eda_qtc_comparison_plot
Description
plots different corrections of QT against RR to compare which to use.
Usage
eda_qtc_comparison_plot( data, rr_col, qt_col, qtcb_col, qtcf_col, qtcp_col = NULL, id_col = NULL, trt_col = NULL, legend_location = "top", model_type = c( "lm", "lme"), show_model_results = TRUE, method = "REML", remove_rr_iiv = FALSE, conf_int = 0.9, style = list())Arguments
| Name | Description |
|---|---|
data | dataframe containing QTc data |
rr_col | an unquoted column name of RR data |
qt_col | an unquoted column name of QT data |
qtcb_col | an unquoted column name of Bazett corrected QT data |
qtcf_col | an unquoted column name of Fridericia corrected QT data |
qtcp_col | Optional - an unquoted column name of Population corrected QT data |
id_col | an unquoted column name of ID data |
trt_col | Optional - an unquoted column name of treatment group data |
legend_location | string for legend location, top,bottom,left,right |
model_type | lm or lme, which model to fit for showing on plot |
show_model_results | a bool for showing regression slope on plot. |
method | method for nlme::lme fitting |
remove_rr_iiv | a boolean for removing IIV on RR slope |
conf_int | confidence interval for lm results default 0.90 |
style | named list of any argument that can be passed to style_plots |
Returns
a plot
Examples
data <- preprocess(data)
eda_qtc_comparison_plot( data, RR, QT, QTCB, QTCF, id_col = ID, trt_col = TRTG, model_type = "lme", show_model_results = TRUE, remove_rr_iiv = TRUE, legend_location = 'top')